Conventions of genetic operator design and registration in geppy¶
To allow maximal flexibility in GEP-based algorithm design, the genetic operators are not hard-coded in geppy. Instead, users can define their own operators for all operations, like selection, mutation and crossover, quite easily while still enjoying the builtin algorithm support. In Python, instead of common interface implementation in languages like Java, we simply prefer duck typing. With regard to the genetic operators, you can define your own operators without implementing an interface explicitly but simply by following some conventions. In this tutorial, the conventions for genetic operator definition and their incorporation into a toolbox are introduced.
Skeleton of an evolutionary algorithm and the registration of operators¶
The following figure shows the simplified flowchart of a typical evolutionary algorithm, for example, the gene expression programming (GEP) algorithm.
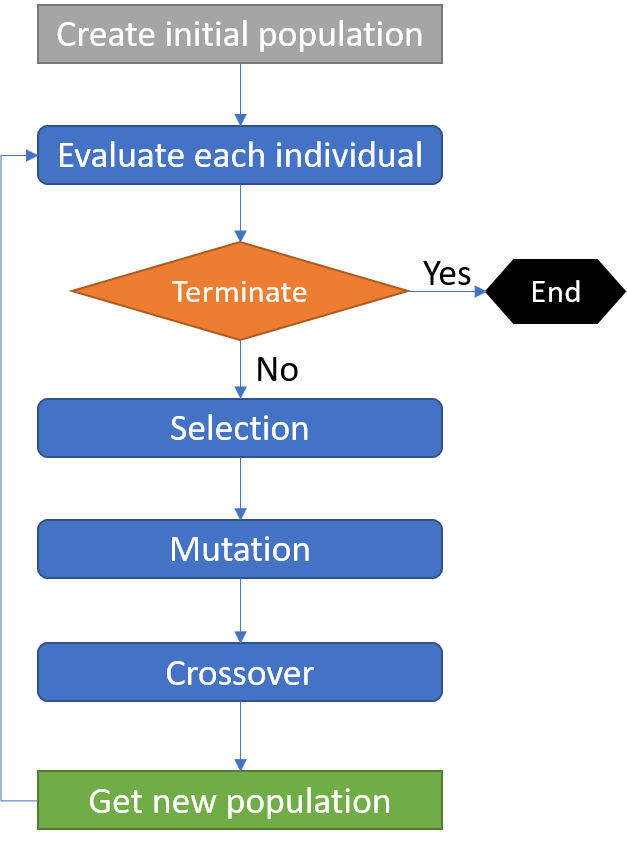
Accordingly, for the basic GEP algorithm, we need to provide at least three classes of operators, i.e., selection, mutation and crossover. As recommended in DEAP, we usually register all the genetic operators into a toolbox and then launch an algorithm by passing the toolbox into it. In such a way, the toolbox can be used to access the operators by predefined names and the probability for each operator is given as arguments of the builtin algorithms. Please check the DEAP documentation on Operators and Algorithms for more details.
However, unlike the traditional genetic algorithm or genetic programming, there are significantly more genetic operators in GEP, which are listed in the detailed flowchart of GEP in the Introduction to gene expression programming tutorial. As a result, if we simply follow the design style of the toolbox and algorithms in DEAP, like deap.algorithms.eaSimple(), the following possible drawbacks are present:
- There will be too many arguments required for an algorithm function to assign the probability to each operator, especially for the GEP-RNC algorithm, where more operators are used.
- It is not flexible enough, because we have to predetermine the names of genetic operators in the toolbox according to the convention of algorithms. For instance, the toolbox.mutate may represent an mutation operator. However, if we have multiple mutation operators in total, then we have to first combine them into a single one by defining a custom operator and then register it into the toolbox with the alias ‘mutate’.
By contrast, in GEP, what we want is:
- We can register as many operators as we like by following a convention with few restrictions.
- The probability of each operator should also be injected into the toolbox instead of specifying them separately as arguments of the builtin algorithm, since the number of operators are not known in advance and there may be too many arguments.
To achieve the above two goals, the following conventions should be respected in geppy when designing your own operators and registering them into the toolbox.
Conventions of genetic operator design¶
As aforementioned, there are significantly more genetic operators in GEP than traditional GA or GP algorithms, including the various transposition operators and crossover operators. However, regarding operators which may modify the individuals, they can be classified into two general types as follows.
- General mutation operator
If an operator accepts a single individual and then returns a modified individual, this operator is considered as a general mutation operator.
In canonical GEP, general mutation operators include point mutation, inversion, IS transposition, RIS transposition and gene transposition. In the GEP-RNC algorithm, the Dc-specific mutation, transposition, inversion and direct RNC-array mutation are also general mutation operators by definition.
- General crossover operator
If an operator accepts two individuals and returns two modified individuals, then this operator is called a general crossover operator.
In canonical GEP, the general crossover operators include one-point recombination and two-point recombination as well as gene recombination.
The above definitions of general mutation/crossover operators are better illustrated with the following figure.
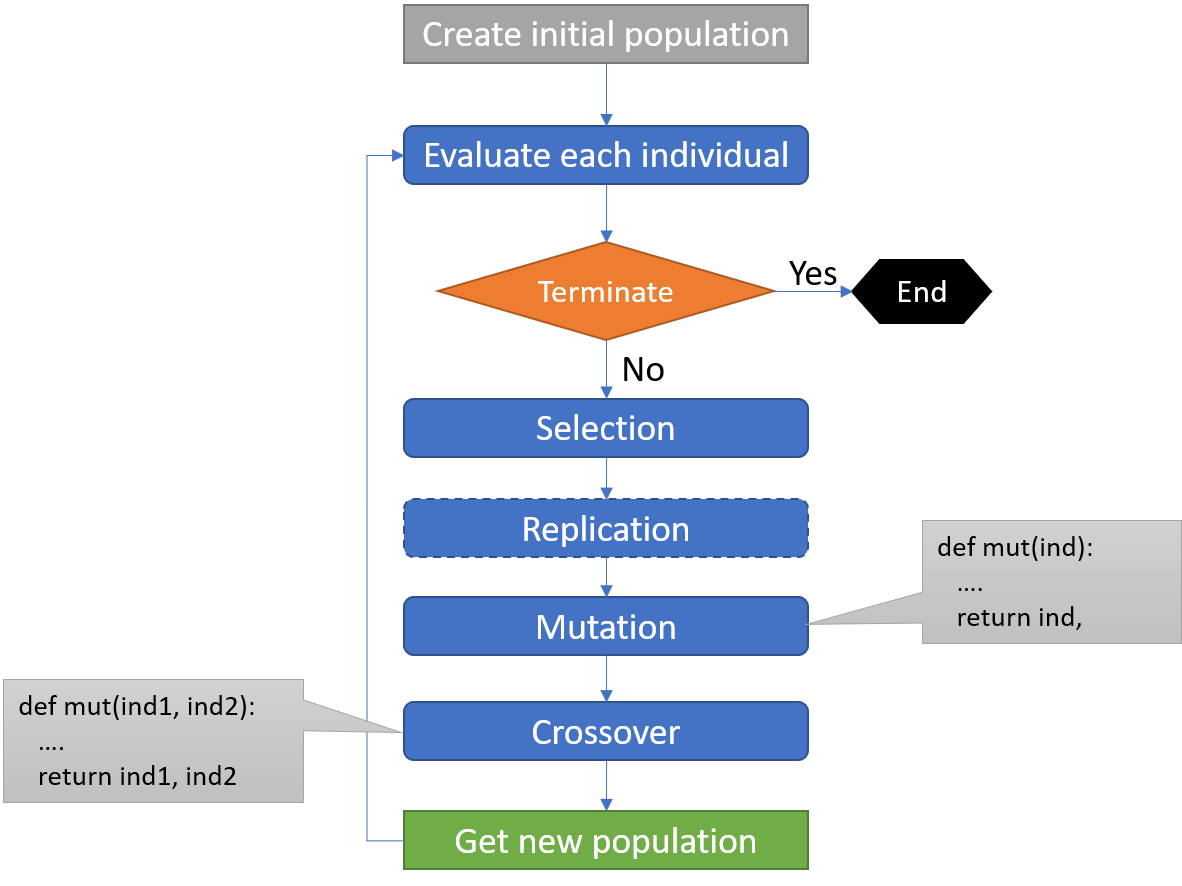
In geppy, the conventions for such operators’ design and registration into a toolbox require the following rules to be obeyed:
- Each toolbox is an instance of the
Toolboxclass. - One and only one selection operator must be registered by the alias select.
- Multiple general mutation operators can be registered, but their aliases in the toolbox must all start with mut.
- Multiple general crossover operators can be registered, but their aliases in the toolbox must all start with cx.
- Each general mutation/crossover operator should have their probability specified in the
pbsproperty with the same alias. Otherwise, this operator is assumed to be assigned a zero probability.
Code example¶
The following code code snippet is extracted from Boolean model symbolic regression showing the registration of genetic operators into a Toolbox
toolbox.register('select', tools.selRoulette)
## general mutations whose aliases start with 'mut'
# We can specify the probability for an operator with the .pbs property
toolbox.register('mut_uniform', gep.mutate_uniform, pset=pset, ind_pb=2 / (2 * h + 1))
toolbox.pbs['mut_uniform'] = 0.1
# Alternatively, assign the probability along with registration using the pb keyword argument.
toolbox.register('mut_invert', gep.invert, pb=0.1)
toolbox.register('mut_is_ts', gep.is_transpose, pb=0.1)
toolbox.register('mut_ris_ts', gep.ris_transpose, pb=0.1)
toolbox.register('mut_gene_ts', gep.gene_transpose, pb=0.1)
## general crossover whose aliases start with 'cx'
toolbox.register('cx_1p', gep.crossover_one_point, pb=0.1)
toolbox.pbs['cx_1p'] = 0.4 # just show that the probability can be overwritten
toolbox.register('cx_2p', gep.crossover_two_point, pb=0.2)
toolbox.register('cx_gene', gep.crossover_gene, pb=0.1)